AnnoQ Help/Tutorial
The backend of the system is a large collection of pre-annotated variants from the Haplotype Reference Consortium (HRC) (~39 million) with sequence features by WGSA and functions by PANTHER and Gene Ontology. The data is built in an Elasticsearch framework and an API was built to allow users to quickly access the annotation data.
This website is free and open to all users and there is no login requirement.
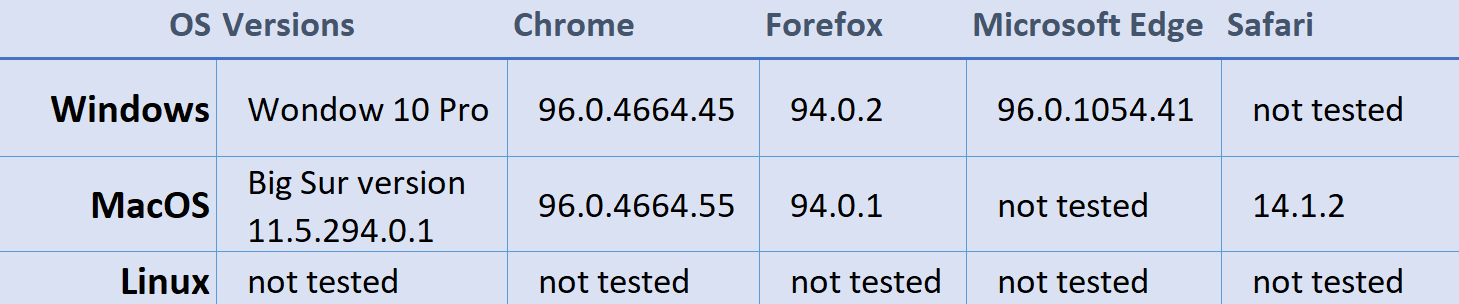
Browser Compatibility Summary
For more details on AnnoQ’s browser support and future work check out further reading
How the documentation is organized
AnnoQ has a lot of documentation. A high-level overview of how it’s organized will help you know where to look for certain things:
AnnoQ Tutorials take you by the hand through a series of steps to use the Interactive Query UI, API and R package. Start here if you’re new to AnnoQ. Also look at the “First steps” below.
Topic guides discuss key topics and concepts at a fairly high level and provide useful background information and explanation.
How-to guides are recipes. They guide you through the steps involved in addressing key problems and use-cases. They are more advanced than tutorials and assume some knowledge of how AnnoQ works.
What can you do with AnnoQ?
- Access large scale genetic variant annotations
- Interactive Query UI: Getting the annotation data with a graphic interface.
- API Data Access: Retrieving annotation data using scripts.
- R Package: Getting annotation data via R programming language.
- Choose a query type which suits you background
- Genome Coordinates Query
- Query by Upload VCF file
- Gene Product Query
- rsID query
- General Keyword Searech
- Easily download the results of annotation data
- csv file
- tsv file
- Export configuration file and use the API
With AnnoQ Interactive Query UI, you can query, advanced queries, upload a VCF file of up to 10,000 variants right in your web browser—no special software is required.
Quick Start
For a Quick Tutorial, check out Quick Tutorial Docs
For release notes, check out Changelog