AnnoQ Help/Tutorial
AnnoQ is a platform that integrates a datastore of pre-annotated SNPs, APIs and packages for accessing the SNPs programmatically and a website for viewing the SNP data. The backend of the system is a large collection of pre-annotated variants from the Haplotype Reference Consortium (HRC) (~39 million) with sequence features by WGSA and functions by PANTHER , Gene Ontology, Reactome and PEREGRINE enhancer mappings. The annotations have also been categorized to allow users easy access to specific subsets of data. The data is built on an Elasticsearch framework which can be accessed via API.
AnnoQ Tutorials gives a high-level overview of the system with details about the Interactive Query UI, API, and software packages for programmatic access.
This website is free and open to all users and there is no login requirement.
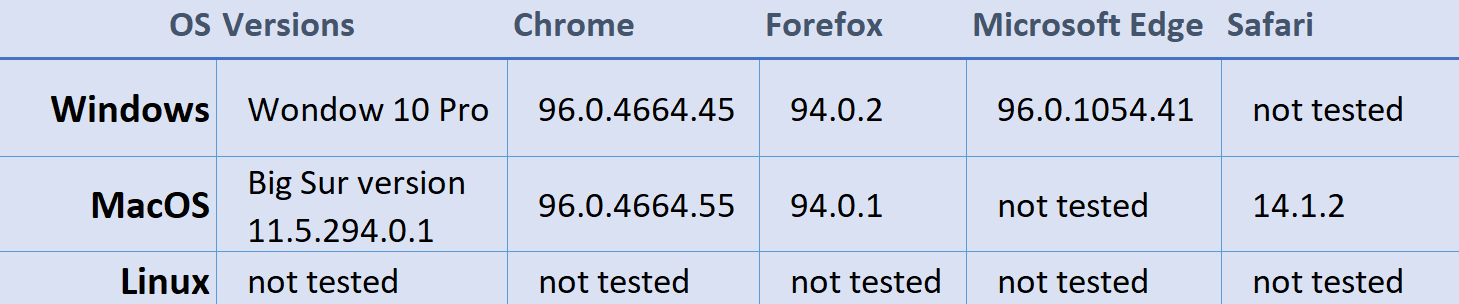
Browser Compatibility Summary
For more details on AnnoQ’s browser support and future work check out further reading
What can you do with AnnoQ?
Access large scale genetic variant annotations
- Interactive Query UI: Retrieve and or view annotation data using a graphical user interface on standard web browsers.
- Search for SNPs using uing:
- Genome Coordinates
- VCF file upload
- Gene Product
- rsID
- Download search results formatted as:
- csv file
- tsv file
- Export configuration file with search fields of interest to use with the API, or for future searches or sharing
- Search for SNPs using uing:
-
AnnoQ Services: Retrieve annotation data from the command line or scripts.
-
R Package: Retrieve annotation data via the R programming language.
- Python Library: Retrieve annotation data using the Python programming language.