Combines two or more MCMC runs into a single run. If runs have multiple chains, it will check that all have the same number of chains, and it will join chains using the rbind function.
append_chains(...)Value
If mcmc.list, an object of class mcmc.list, otherwise,
an object of class mcmc.
Examples
# Appending two chains
data("lifeexpect")
logpost <- function(p) {
sum(with(lifeexpect, dnorm(
age - p[1] - smoke * p[2] - female * p[3],
sd = p[4], log = TRUE)
))
}
# Using the RAM kernel
kern <- kernel_ram(lb = c(-100, -100, -100, .00001))
init <- c(
avg_age = 70,
smoke = 0,
female = 0,
sd = 1
)
ans0 <- MCMC(initial = init, fun = logpost, nsteps = 1000, seed = 22, kernel = kern)
ans1 <- MCMC(initial = ans0, fun = logpost, nsteps = 2000, seed = 55, kernel = kern)
ans2 <- MCMC(initial = ans1, fun = logpost, nsteps = 2000, seed = 1155, kernel = kern)
ans_tot <- append_chains(ans0, ans1, ans2)
# Looking at the posterior distributions (see ?lifeexpect for info about
# the model). Only the trace
op <- par(mfrow = c(2,2))
for (i in 1:4)
coda::traceplot(ans_tot[, i, drop=FALSE])
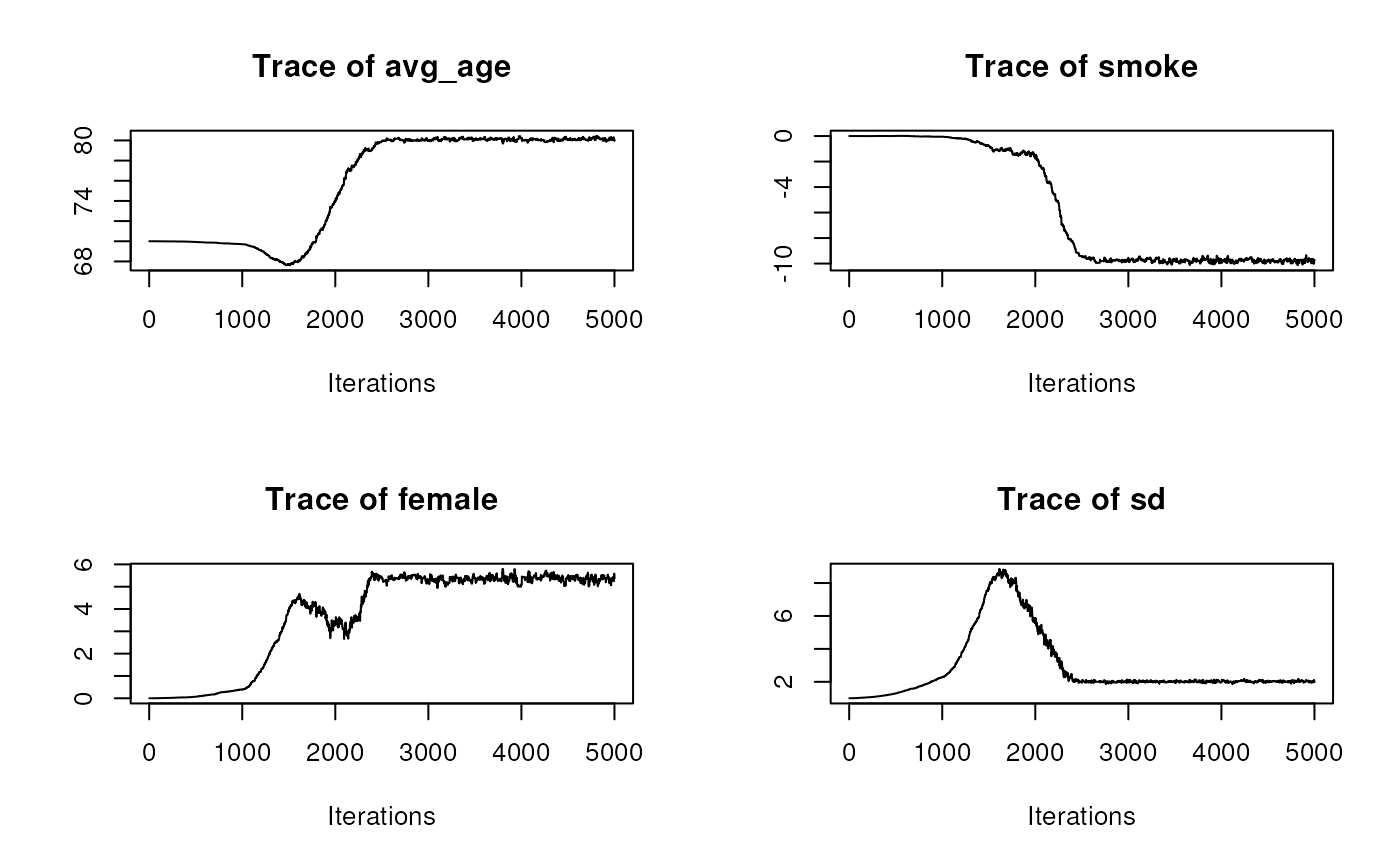 par(op)
par(op)