The function is a wrapper of stats::optim().
Arguments
- model
A model as specified in aphylo-model.
- params
A vector of length 7 with initial parameters. In particular
psi[1],psi[2],mu[1],mu[2],eta[1],eta[2]andPi.- method, control, lower, upper
Arguments passed to
stats::optim().- priors
A function to be used as prior for the model (see bprior).
- check_informative
Logical scalar. When
TRUEthe algorithm stops with an error when the annotations are uninformative (either 0s or 1s).- reduced_pseq
Logical. When
TRUEit will use a reduced peeling sequence in which it drops unannotated leafs. If the model includesetathis is set toFALSE.
Value
An object of class aphylo_estimates.
Details
The default starting parameters are described in APHYLO_PARAM_DEFAULT.
See also
Other parameter estimation:
APHYLO_DEFAULT_MCMC_CONTROL
Examples
# Using simulated data ------------------------------------------------------
set.seed(19)
dat <- raphylo(100)
dat <- rdrop_annotations(dat, .4)
# Computing Estimating the parameters
ans <- aphylo_mle(dat ~ psi + mu_d + eta + Pi)
ans
#> Warning: NaNs produced
#>
#> ESTIMATION OF ANNOTATED PHYLOGENETIC TREE
#>
#> Call: aphylo_mle(model = dat ~ psi + mu_d + eta + Pi)
#> LogLik: -35.6771
#> Method used: L-BFGS-B (20 steps)
#> convergence: 0 (see ?optim)
#> # of Leafs: 100
#> # of Functions 1
#> # of Trees: 1
#>
#> Estimate Std. Err.
#> psi0 0.0000 NaN
#> psi1 0.0000 0.4375
#> mu_d0 1.0000 0.6516
#> mu_d1 0.4259 0.4224
#> eta0 1.0000 0.2357
#> eta1 1.0000 0.1543
#> Pi 1.0000 3.2408
#>
# Plotting the path
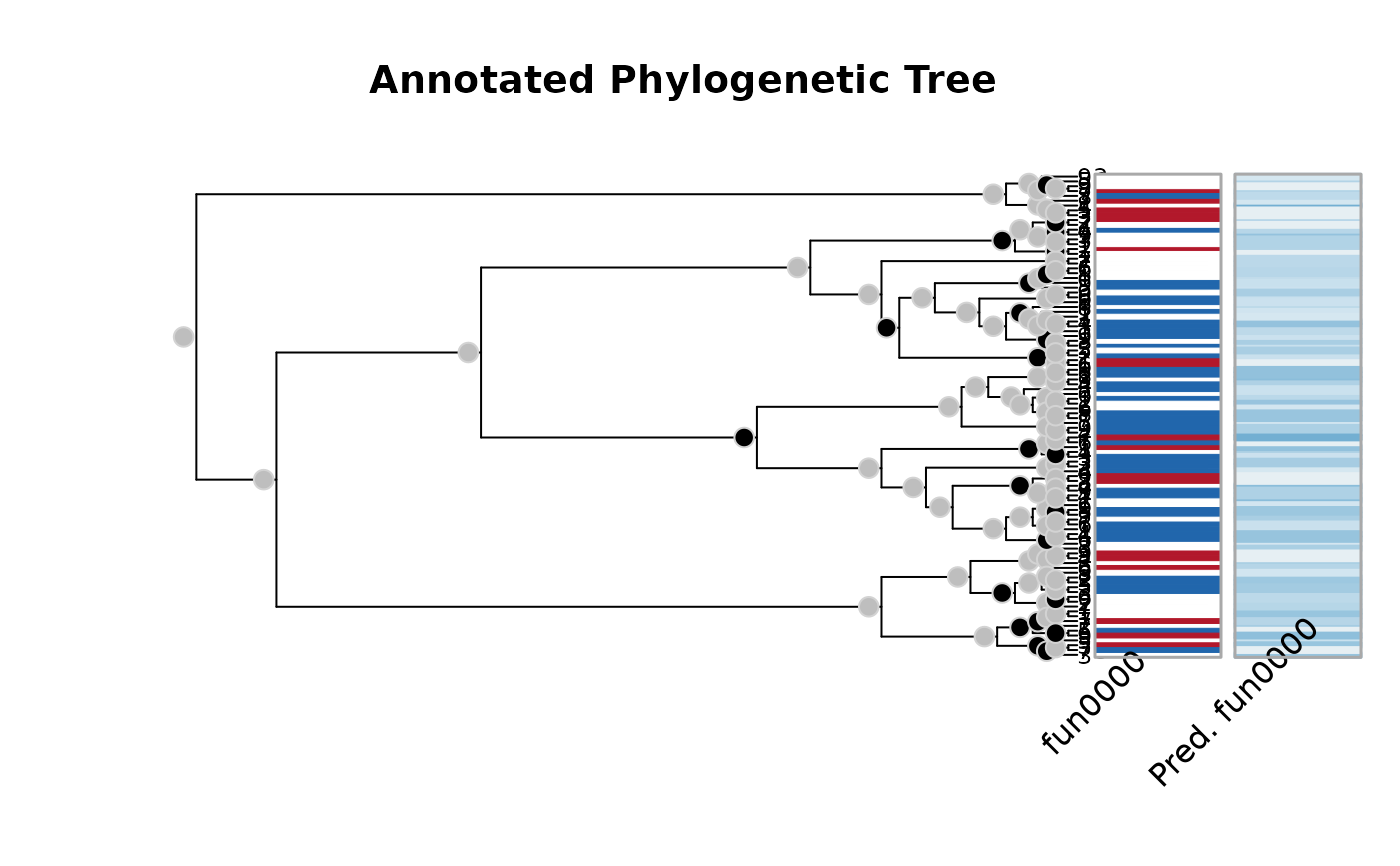
plot(ans)
 # Computing Estimating the parameters Using Priors for all the parameters
mypriors <- function(params) {
dbeta(params, c(2, 2, 2, 2, 1, 10, 2), rep(10, 7))
}
ans_dbeta <- aphylo_mle(dat ~ psi + mu_d + eta + Pi, priors = mypriors)
ans_dbeta
#>
#> ESTIMATION OF ANNOTATED PHYLOGENETIC TREE
#>
#> Call: aphylo_mle(model = dat ~ psi + mu_d + eta + Pi, priors = mypriors)
#> LogLik (unnormalized): -50.2767
#> Method used: L-BFGS-B (29 steps)
#> convergence: 0 (see ?optim)
#> # of Leafs: 100
#> # of Functions 1
#> # of Trees: 1
#>
#> Estimate Std. Err.
#> psi0 0.1144 0.0896
#> psi1 0.0362 0.0371
#> mu_d0 0.1370 0.0554
#> mu_d1 0.0422 0.0288
#> eta0 0.6666 0.0907
#> eta1 0.8500 0.0461
#> Pi 0.0910 0.0868
#>
# Computing Estimating the parameters Using Priors for all the parameters
mypriors <- function(params) {
dbeta(params, c(2, 2, 2, 2, 1, 10, 2), rep(10, 7))
}
ans_dbeta <- aphylo_mle(dat ~ psi + mu_d + eta + Pi, priors = mypriors)
ans_dbeta
#>
#> ESTIMATION OF ANNOTATED PHYLOGENETIC TREE
#>
#> Call: aphylo_mle(model = dat ~ psi + mu_d + eta + Pi, priors = mypriors)
#> LogLik (unnormalized): -50.2767
#> Method used: L-BFGS-B (29 steps)
#> convergence: 0 (see ?optim)
#> # of Leafs: 100
#> # of Functions 1
#> # of Trees: 1
#>
#> Estimate Std. Err.
#> psi0 0.1144 0.0896
#> psi1 0.0362 0.0371
#> mu_d0 0.1370 0.0554
#> mu_d1 0.0422 0.0288
#> eta0 0.6666 0.0907
#> eta1 0.8500 0.0461
#> Pi 0.0910 0.0868
#>