The model fitting of annotated phylogenetic trees can be done using either
MLE via aphylo_mle() or MCMC via aphylo_mcmc(). This section describes
the object of class aphylo_estimates that these functions generate and
the post estimation methods/functions that can be used.
# S3 method for class 'aphylo_estimates'
print(x, ...)
# S3 method for class 'aphylo_estimates'
coef(object, ...)
# S3 method for class 'aphylo_estimates'
vcov(object, ...)
# S3 method for class 'aphylo_estimates'
plot(
x,
y = NULL,
which.tree = 1L,
ids = list(1:Ntip(x)[which.tree]),
loo = TRUE,
nsamples = 1L,
ncores = 1L,
centiles = c(0.025, 0.5, 0.975),
cl = NULL,
...
)Arguments
- x, object
Depending of the method, an object of class
aphylo_estimates.- ...
Further arguments passed to the corresponding method.
- y
Ignored.
- which.tree
Integer scalar. Which tree to plot.
- ids, nsamples, ncores, centiles, cl
passed to
predict.aphylo_estimates()- loo
Logical scalar. When
loo = TRUE, predictions are preformed similar to what a leave-one-out cross-validation scheme would be done (see predict.aphylo_estimates).
Value
Objects of class aphylo_estimates are a list withh the following elements:
- par
A numeric vector of length 5 with the solution.
- hist
A numeric matrix of size
counts*5with the solution path (length 2 if usedoptimas the intermediate steps are not available to the user). In the case ofaphylo_mcmc,histis an object of classcoda::mcmc.list().- ll
A numeric scalar with the value of
fun(par, dat). The value of the log likelihood.- counts
Integer scalar number of steps/batch performed.
- convergence
Integer scalar. Equal to 0 if
optimconverged. Seeoptim.- message
Character scalar. See
optim.- fun
A function (the objective function).
- priors
If specified, the function
priorspassed to the method.- dat
The data
datprovided to the function.- par0
A numeric vector of length 5 with the initial parameters.
- method
Character scalar with the name of the method used.
- varcovar
A matrix of size 5*5. The estimated covariance matrix.
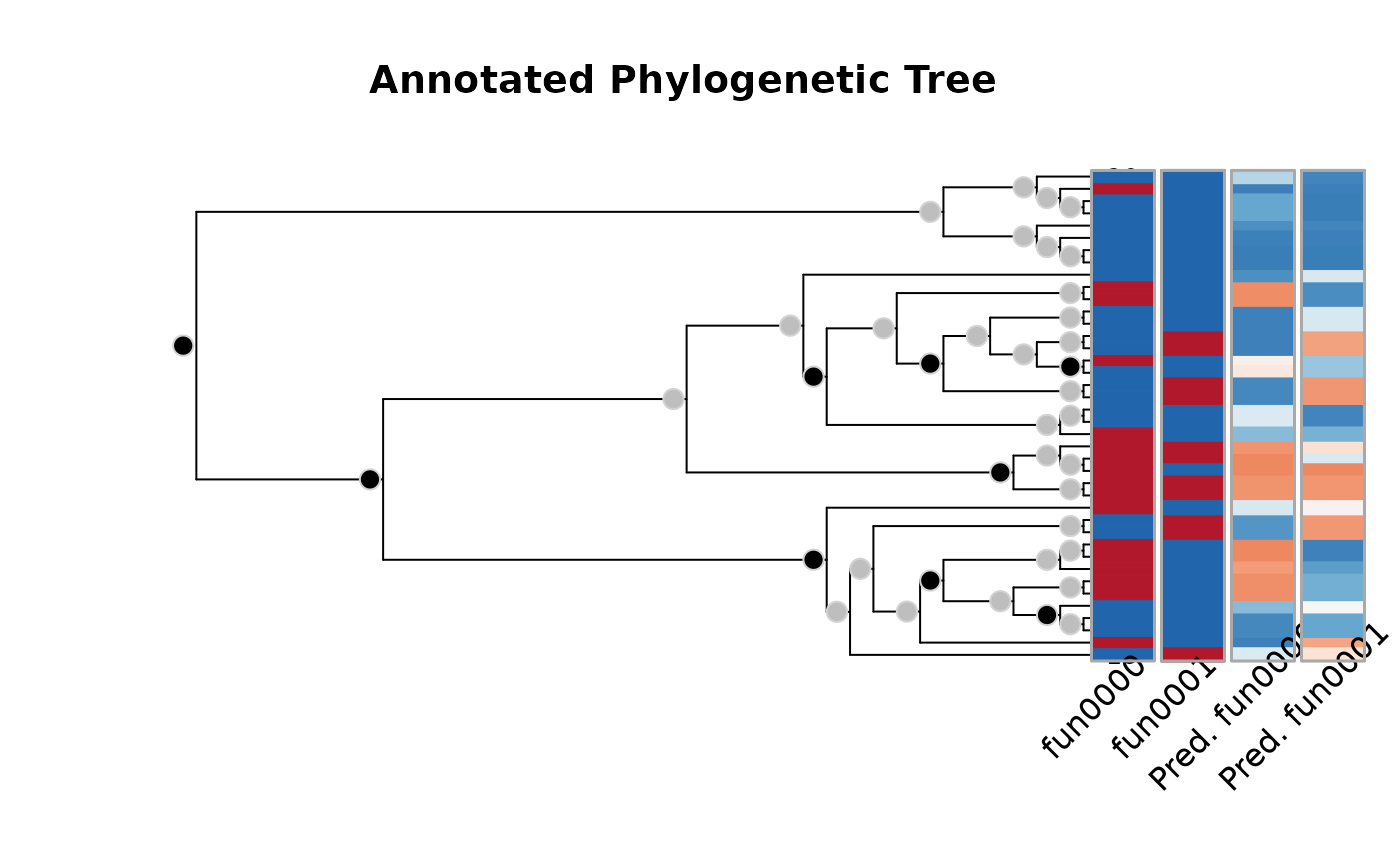
The plot method for aphylo_estimates returns the selected tree
(which.tree) with predicted annotations, also of class aphylo.
Details
The plot method for the object of class aphylo_estimates plots
the original tree with the predicted annotations.
Examples
set.seed(7881)
atree <- raphylo(40, P = 2)
res <- aphylo_mcmc(atree ~ mu_d + mu_s + Pi)
#> Warning: While using multiple chains, a single initial point has been passed via `initial`: c(0.9, 0.5, 0.1, 0.05, 0.5). The values will be recycled. Ideally you would want to start each chain from different locations.
#> Convergence has been reached with 10000 steps. Gelman-Rubin's R: 1.0233. (500 final count of samples).
print(res)
#>
#> ESTIMATION OF ANNOTATED PHYLOGENETIC TREE
#>
#> Call: aphylo_mcmc(model = atree ~ mu_d + mu_s + Pi)
#> LogLik: -37.7731
#> Method used: mcmc (10000 steps)
#> # of Leafs: 40
#> # of Functions 2
#> # of Trees: 1
#>
#> Estimate Std. Err.
#> mu_d0 0.8392 0.1475
#> mu_d1 0.5985 0.1571
#> mu_s0 0.1515 0.0702
#> mu_s1 0.0544 0.0320
#> Pi 0.5076 0.2971
#>
coef(res)
#> mu_d0 mu_d1 mu_s0 mu_s1 Pi
#> 0.83921480 0.59853604 0.15148002 0.05436977 0.50762014
vcov(res)
#> mu_d0 mu_d1 mu_s0 mu_s1 Pi
#> mu_d0 0.021761402 0.0064170422 -0.0013305666 -0.0000281820 0.0035925925
#> mu_d1 0.006417042 0.0246873308 0.0027730753 -0.0007584415 -0.0038793095
#> mu_s0 -0.001330567 0.0027730753 0.0049245626 -0.0003601076 0.0014041022
#> mu_s1 -0.000028182 -0.0007584415 -0.0003601076 0.0010221978 -0.0001894985
#> Pi 0.003592593 -0.0038793095 0.0014041022 -0.0001894985 0.0882936562
plot(res)